Software
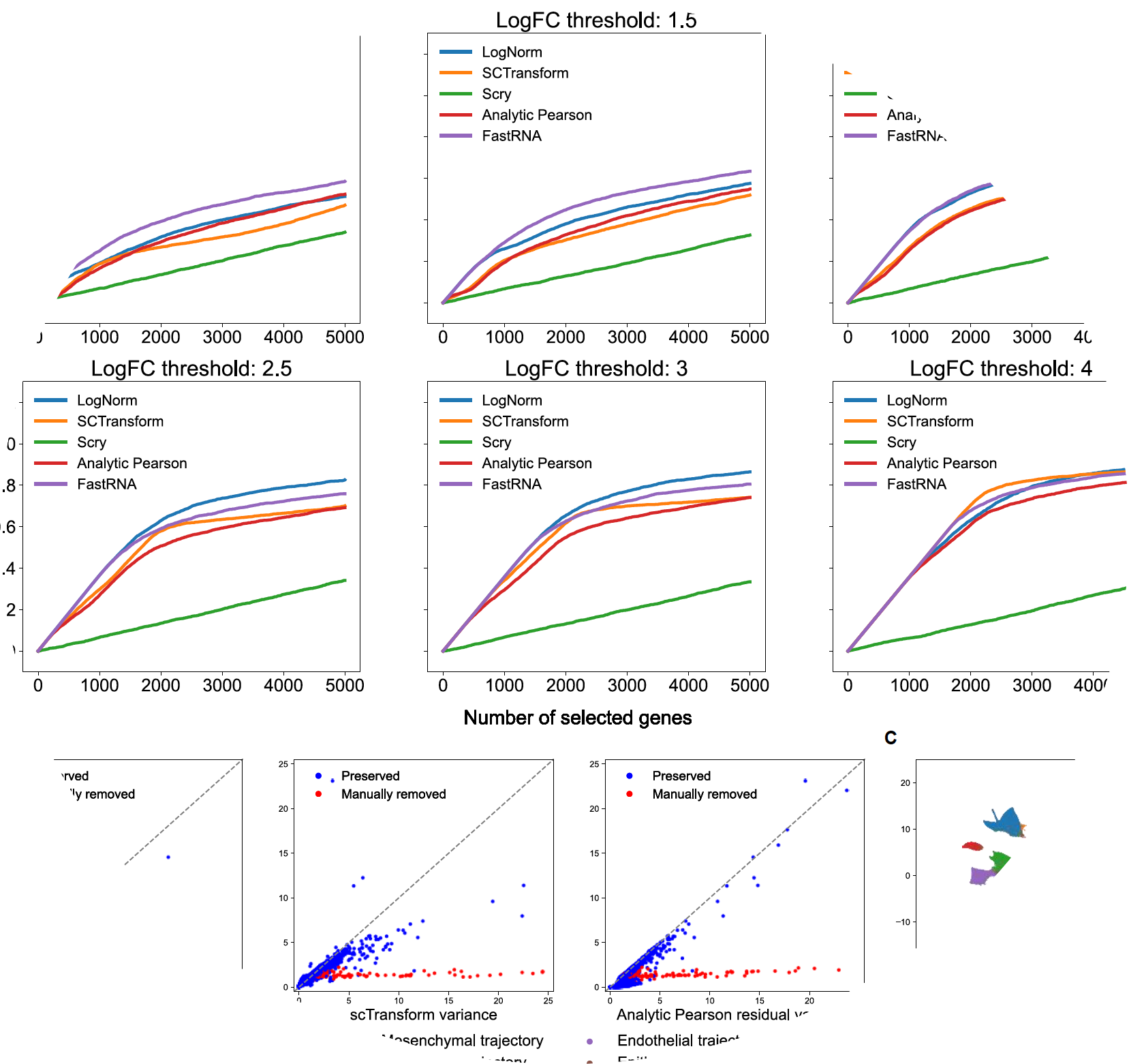
fastRNA is a scalable framework for single-cell RNA sequencing (scRNA-seq) analysis.
fastRNA is a scalable framework for single-cell RNA sequencing (scRNA-seq) analysis.
Hanbin Lee, and Buhm Han. “FastRNA: an efficient solution for PCA of single-cell RNA sequencing data based on a batch-accounting count model.” Am J Hum Genet, in press (2022).
Keyword : #single-cell
Hanbin Lee, and Buhm Han. “FastRNA: an efficient solution for PCA of single-cell RNA sequencing data based on a batch-accounting count model.” Am J Hum Genet, in press (2022).
Keyword : #single-cell
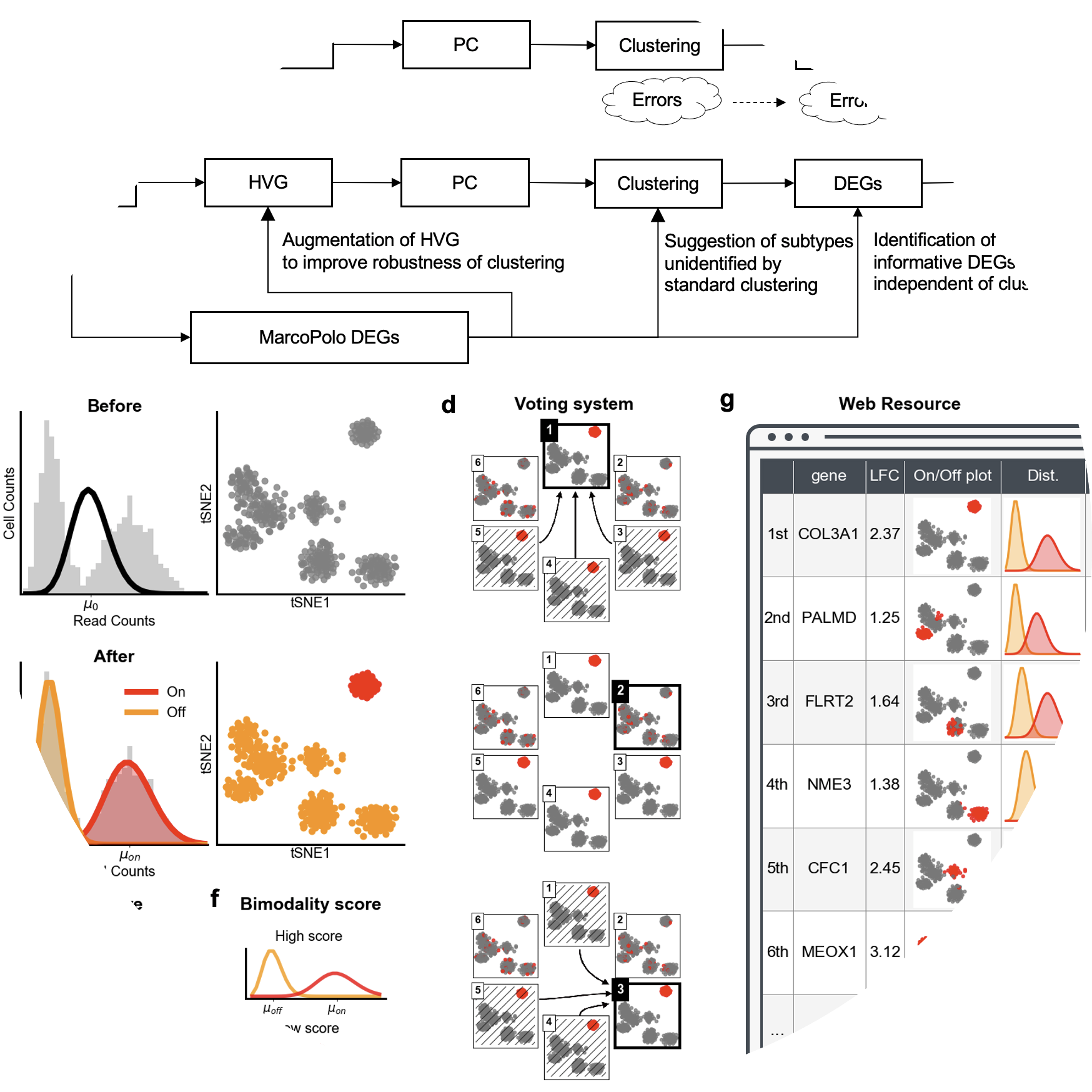
MarcoPolo is a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering
MarcoPolo is a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering
Kim, Chanwoo, Hanbin Lee, Juhee Jeong, Keehoon Jung, and Buhm Han. “MarcoPolo: a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering.” Nucleic Acids Research (2022).
Keyword : #single-cell
Kim, Chanwoo, Hanbin Lee, Juhee Jeong, Keehoon Jung, and Buhm Han. “MarcoPolo: a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering.” Nucleic Acids Research (2022).
Keyword : #single-cell
GAT is a generic tool for conducting GWAS(Genome-Wide Association Study) on phased/unphased(=dosage) biallelic/multiallelic markers simultaneously.
GAT is a generic tool for conducting GWAS(Genome-Wide Association Study) on phased/unphased(=dosage) biallelic/multiallelic markers simultaneously.
Chanwoo Kim, Young Jin Kim, Wanson Choi, Hye-Mi Jang, Mi Yeong Hwang, Sunwoo Jung, Hyunjoon Lim, Sang Bin Hong, Kyungheon Yoon, Bong-Jo Kim, Hyun-Young Park, Buhm Han. "Phenome-wide association study of the major histocompatibility complex region in the Korean population identifies novel association signals." Human Molecular Genetics, 31(15):2655-2667. 2022
Keyword: #GWAS, #PheWAS, #HLA
Chanwoo Kim, Young Jin Kim, Wanson Choi, Hye-Mi Jang, Mi Yeong Hwang, Sunwoo Jung, Hyunjoon Lim, Sang Bin Hong, Kyungheon Yoon, Bong-Jo Kim, Hyun-Young Park, Buhm Han. "Phenome-wide association study of the major histocompatibility complex region in the Korean population identifies novel association signals." Human Molecular Genetics, 31(15):2655-2667. 2022
Keyword: #GWAS, #PheWAS, #HLA
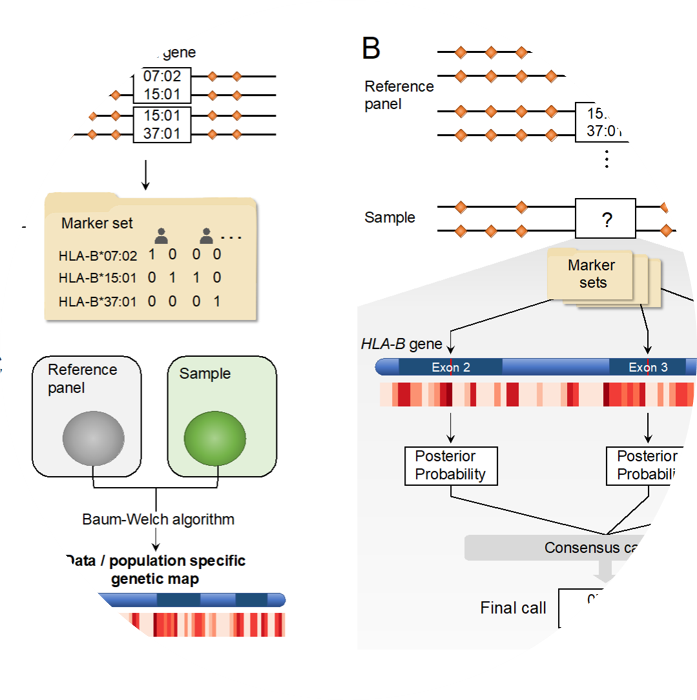
CookHLA imputes HLA genes and amino acids from SNP genotype data.
CookHLA imputes HLA genes and amino acids from SNP genotype data.
S. Cook, W. Choi, H. Lim, Y. Luo, K. Kim, X. Jia, S. Raychaudhuri and Buhm Han, “CookHLA: Accurate Imputation of Human Leukocyte Antigens”. Nature Communications. 2021.
Keyword : #HLA, #imputation, #snp2hla
S. Cook, W. Choi, H. Lim, Y. Luo, K. Kim, X. Jia, S. Raychaudhuri and Buhm Han, “CookHLA: Accurate Imputation of Human Leukocyte Antigens”. Nature Communications. 2021.
Keyword : #HLA, #imputation, #snp2hla
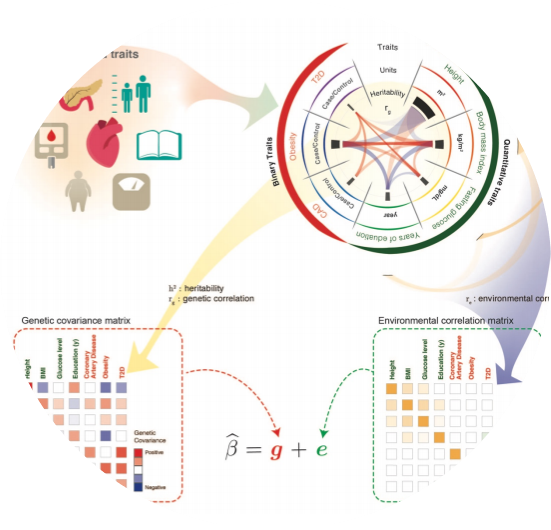
PLEIO offers a summary-statistic-based framework to map and interpret pleiotropic loci in a joint analysis of multiple traits.
PLEIO offers a summary-statistic-based framework to map and interpret pleiotropic loci in a joint analysis of multiple traits.
Lee, Cue Hyunkyu, Huwenbo Shi, Bogdan Pasaniuc, Eleazar Eskin, and Buhm Han. “PLEIO: a method to map and interpret pleiotropic loci with GWAS summary statistics.” The American Journal of Human Genetics 108, no. 1 (2021): 36-48.
Keyword : #pleiotropic, #meta-analysis
Lee, Cue Hyunkyu, Huwenbo Shi, Bogdan Pasaniuc, Eleazar Eskin, and Buhm Han. “PLEIO: a method to map and interpret pleiotropic loci with GWAS summary statistics.” The American Journal of Human Genetics 108, no. 1 (2021): 36-48.
Keyword : #pleiotropic, #meta-analysis
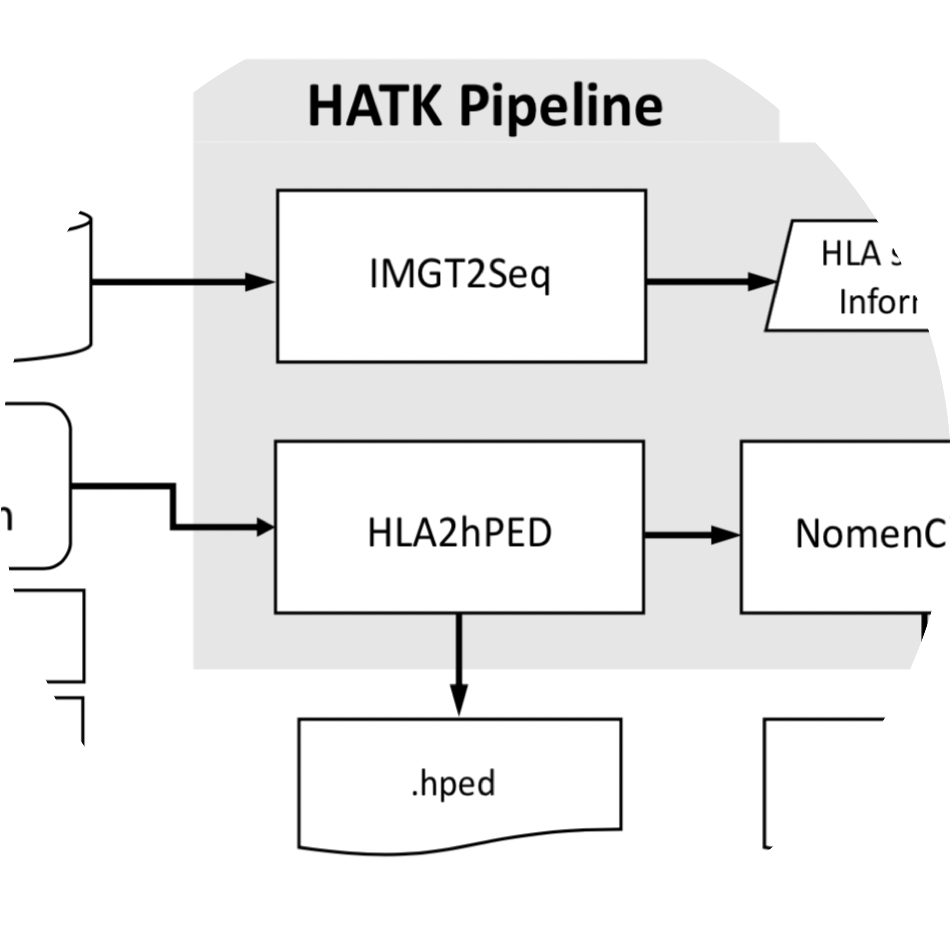
HATK imputes HLA genes and amino acids from SNP genotype data.
HATK imputes HLA genes and amino acids from SNP genotype data.
Wanson Choi, Yang Luo, Soumya Raychaudhuri, Buhm Han. “HATK:HLA-Analysis Tool-kit” Bioinformatics. 2020.
Keyword : #HLA, #imputation
Wanson Choi, Yang Luo, Soumya Raychaudhuri, Buhm Han. “HATK:HLA-Analysis Tool-kit” Bioinformatics. 2020.
Keyword : #HLA, #imputation