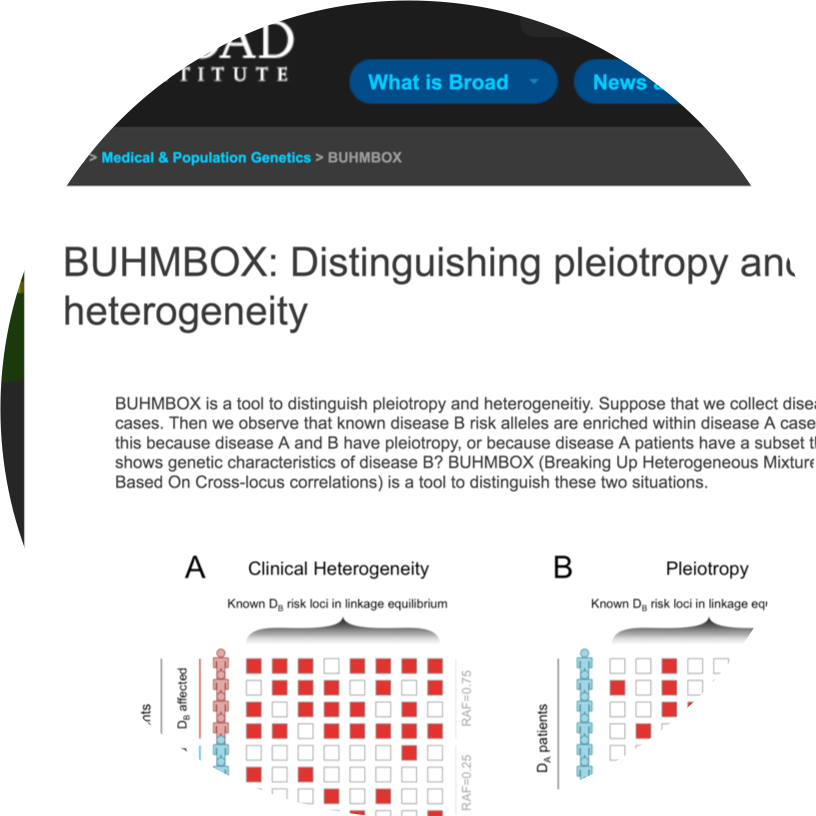
BUHMBOX
BUHMBOX is a tool to distinguish pleiotropy and clinical heterogeneity.
- Buhm Han, Jennie G Pouget, Kamil Slowikowski, Eli Stahl, Cue Hyunkyu Lee, Dorothee Diogo, Xinli Hu, Yu Rang Park, Eunji Kim, Peter K Gregersen, Solbritt Rantapaa Dahqvist, Jane Worthington, Steve Eyre, Lars Klareskog, Tom Huizinga, Wei-Min Chen, Suna Onengut-Gumuscu, Stephen S Rich, Major Depressive Disorder Working Group of the PGC, Naomi Wray, Soumya Raychaudhuri “A method to decipher pleiotropy by detecting underlying heterogeneity driven by hidden subgroups applied to autoimmune and neuropsychiatric diseases.” Nature Genetics. 48(7):803-10. 2016
- Keyword : #pleiotropy, #heterogeneity, #ng
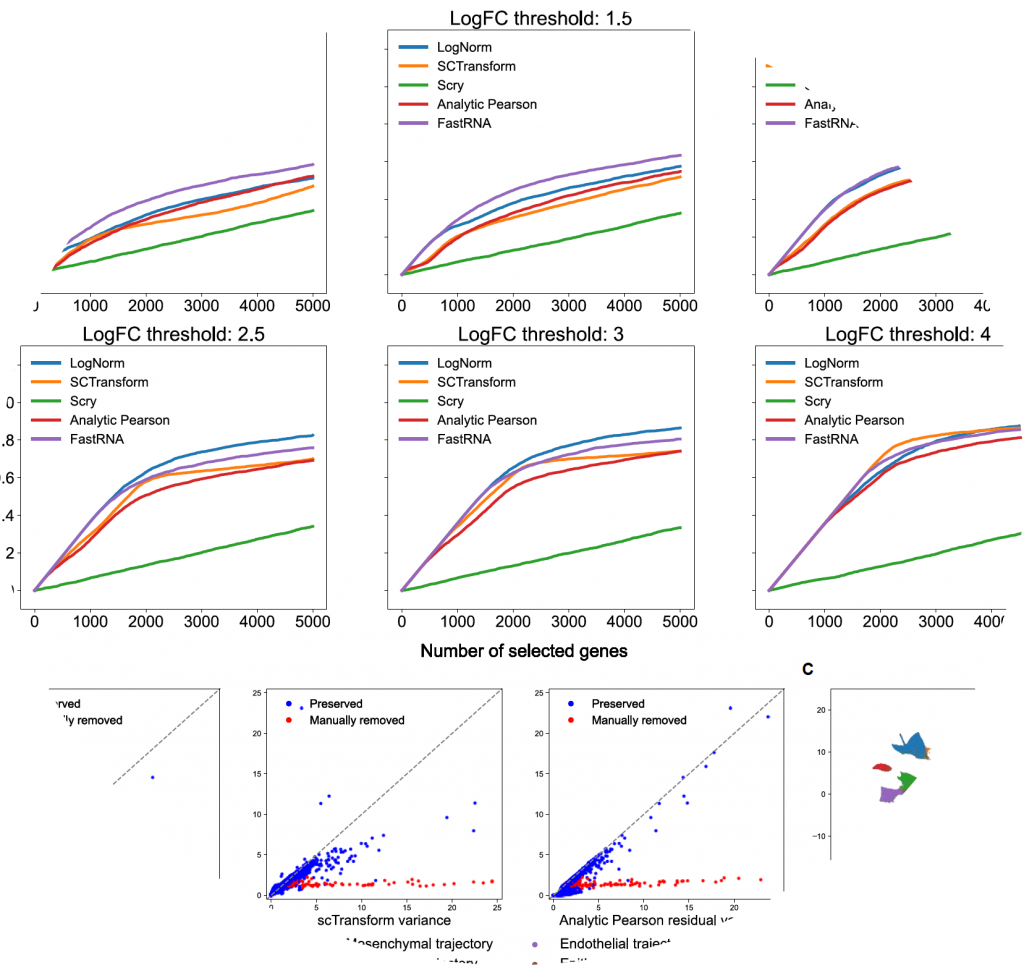
fastRNA
fastRNA is a scalable framework for single-cell RNA sequencing (scRNA-seq) analysis.
- Hanbin Lee, and Buhm Han. “FastRNA: an efficient solution for PCA of single-cell RNA sequencing data based on a batch-accounting count model.” Am J Hum Genet, in press (2022).
- Keyword : #single-cell
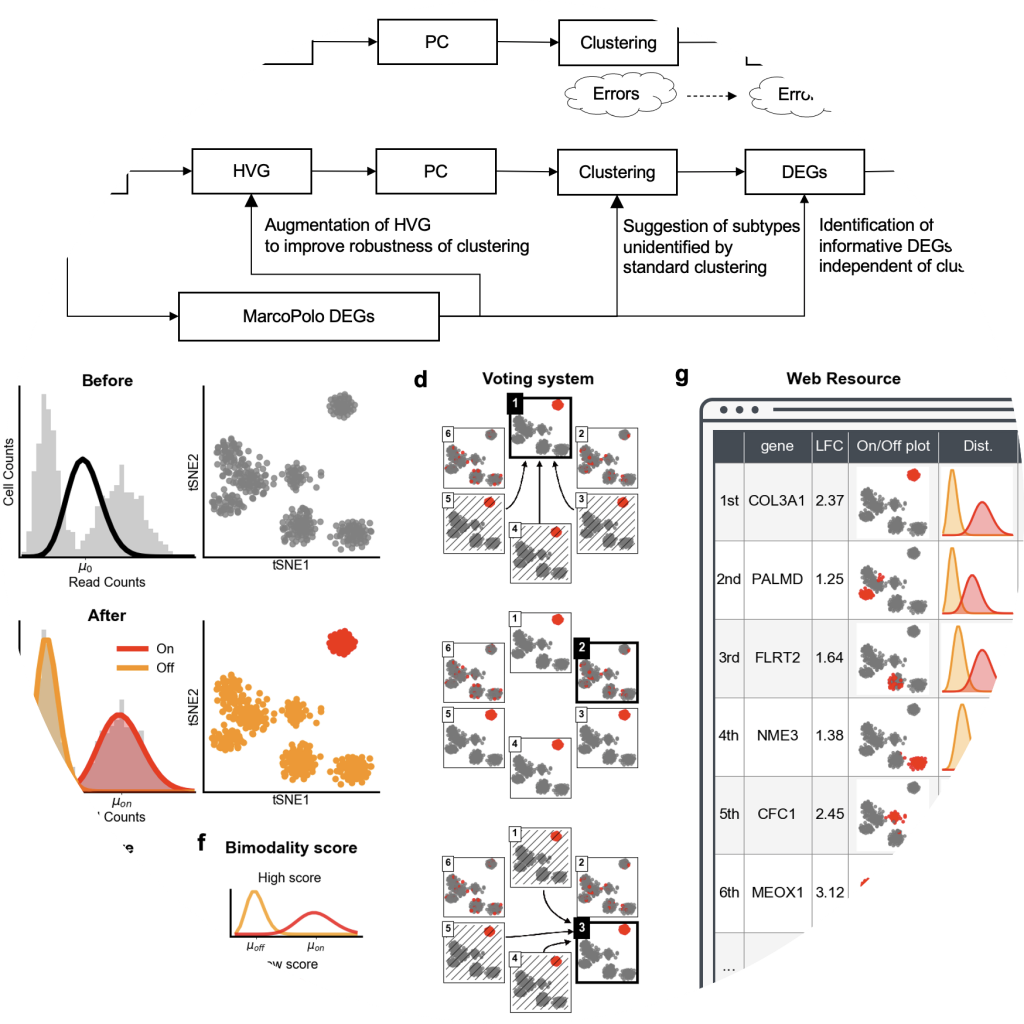
MarcoPolo
MarcoPolo is a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering
- Kim, Chanwoo, Hanbin Lee, Juhee Jeong, Keehoon Jung, and Buhm Han. “MarcoPolo: a method to discover differentially expressed genes in single-cell RNA-seq data without depending on prior clustering.” Nucleic Acids Research (2022).
- Keyword : #single-cell
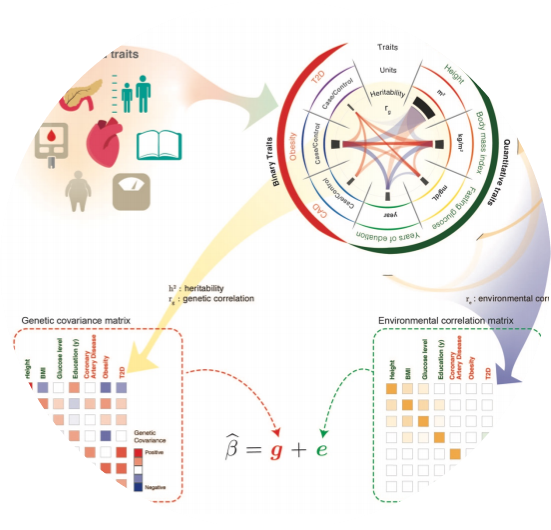
PLEIO
PLEIO offers a summary-statistic-based framework to map and interpret pleiotropic loci in a joint analysis of multiple traits.
- Lee, Cue Hyunkyu, Huwenbo Shi, Bogdan Pasaniuc, Eleazar Eskin, and Buhm Han. “PLEIO: a method to map and interpret pleiotropic loci with GWAS summary statistics.” The American Journal of Human Genetics 108, no. 1 (2021): 36-48.
- Keyword : #pleiotropic, #meta-analysis
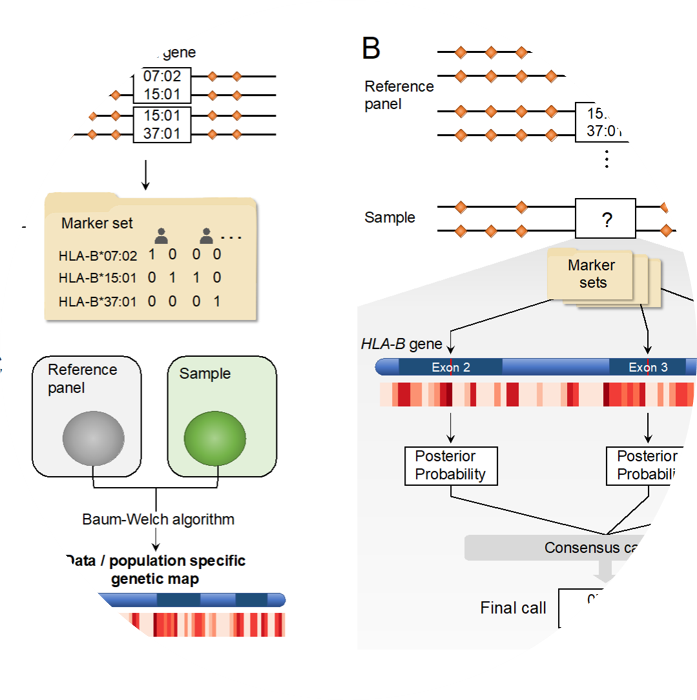
CookHLA
CookHLA imputes HLA genes and amino acids from SNP genotype data.
- S. Cook, W. Choi, H. Lim, Y. Luo, K. Kim, X. Jia, S. Raychaudhuri and Buhm Han, “CookHLA: Accurate Imputation of Human Leukocyte Antigens”. Nature Communications. 2021.
- Keyword : #HLA, #imputation, #snp2hla
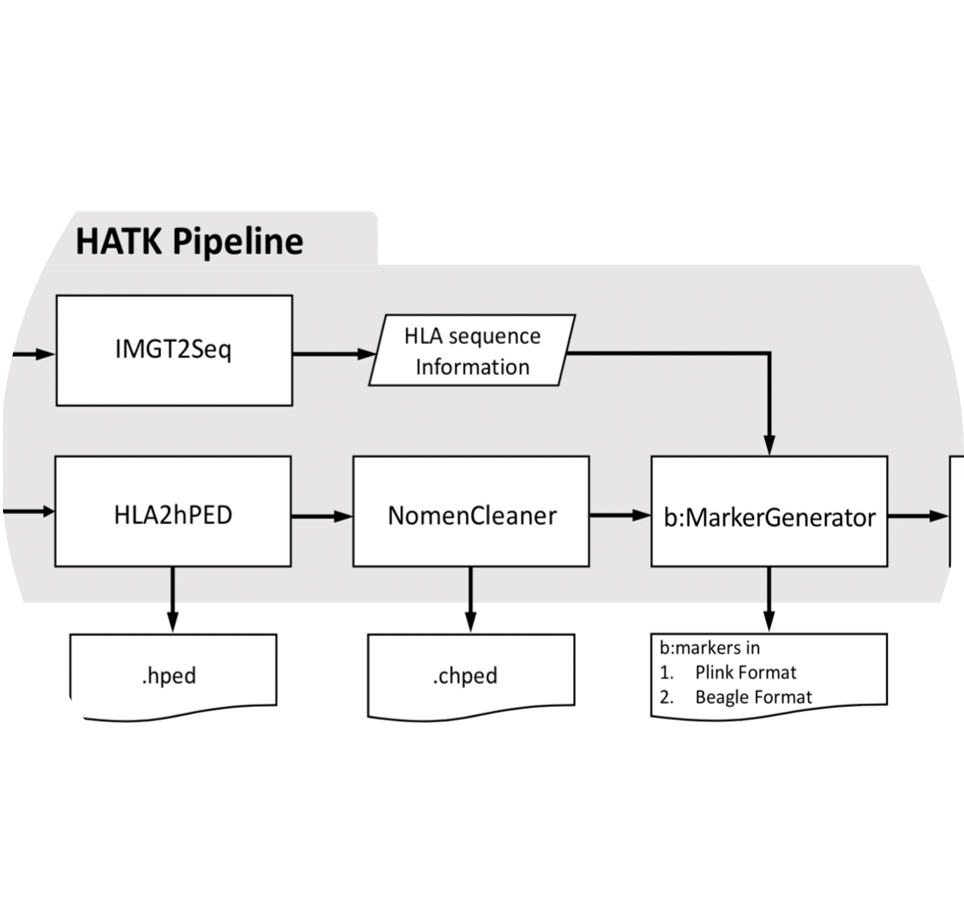
HATK
HATK imputes HLA genes and amino acids from SNP genotype data.
- Wanson Choi, Yang Luo, Soumya Raychaudhuri, Buhm Han. “HATK:HLA-Analysis Tool-kit” Bioinformatics. 2020.
- Keyword : #HLA, #imputation
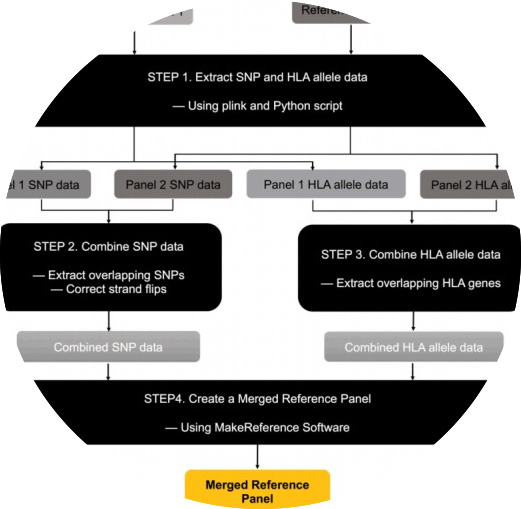
mergeReference
MergeReference merges multiple reference panels for HLA imputation. The merged reference panel could be used for running CookHLA and SNP2HLA.
- Cook, Seungho, and Buhm Han. “MergeReference: a tool for merging reference panels for HLA imputation.” Genomics & Informatics 15, no. 3 (2017): 108.
- Keyword : #HLA, #imputation, #mergereference
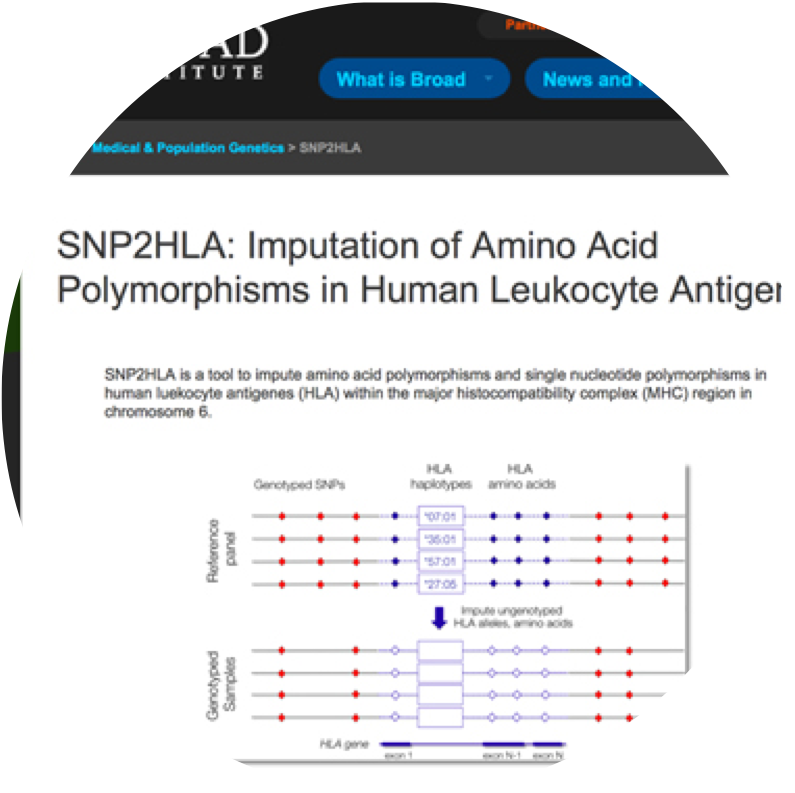
SNP2HLA
SNP2HLA imputes HLA genes and amino acids from SNP genotype data.
- Xiaoming Jia, Buhm Han, Suna Onengut-Gumuscu, Wei-Min Chen, Patrick J. Concannon, Stephen S. Rich, Soumya Raychaudhuri, Paul I.W. de Bakker. “ Imputing Amino Acid Polymorphisms in Human Leukocyte Antigenes. ” PLoS One. 8(6):e64683. 2013.
- Keyword : #HLA, #imputation, #ng
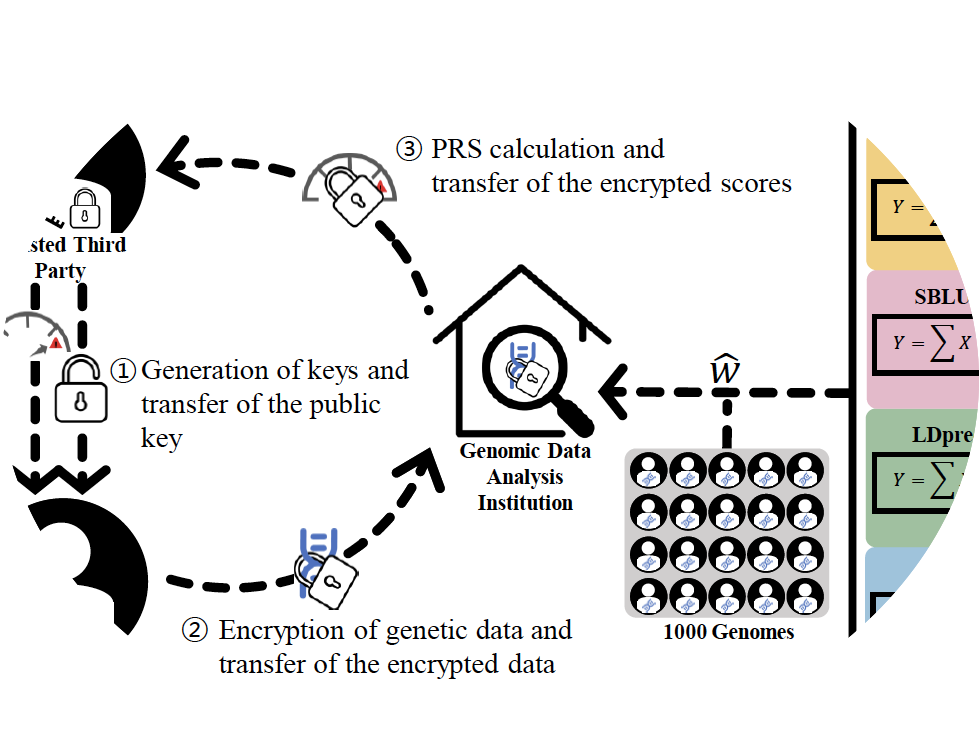
privatePRS
privatePRS is a software tool for polygenic risk scoring, ensuring the privacy issue that each individual can be uniquely identified by genotype data consisting of SNPs.
- Hakin Kim, Buhm Han “ PrivatePRS: privacy-protecting polygenic risk score calculation by homomorphic encryption” Under Review.
- Keyword : #privacy, #homomorphic, #encryption
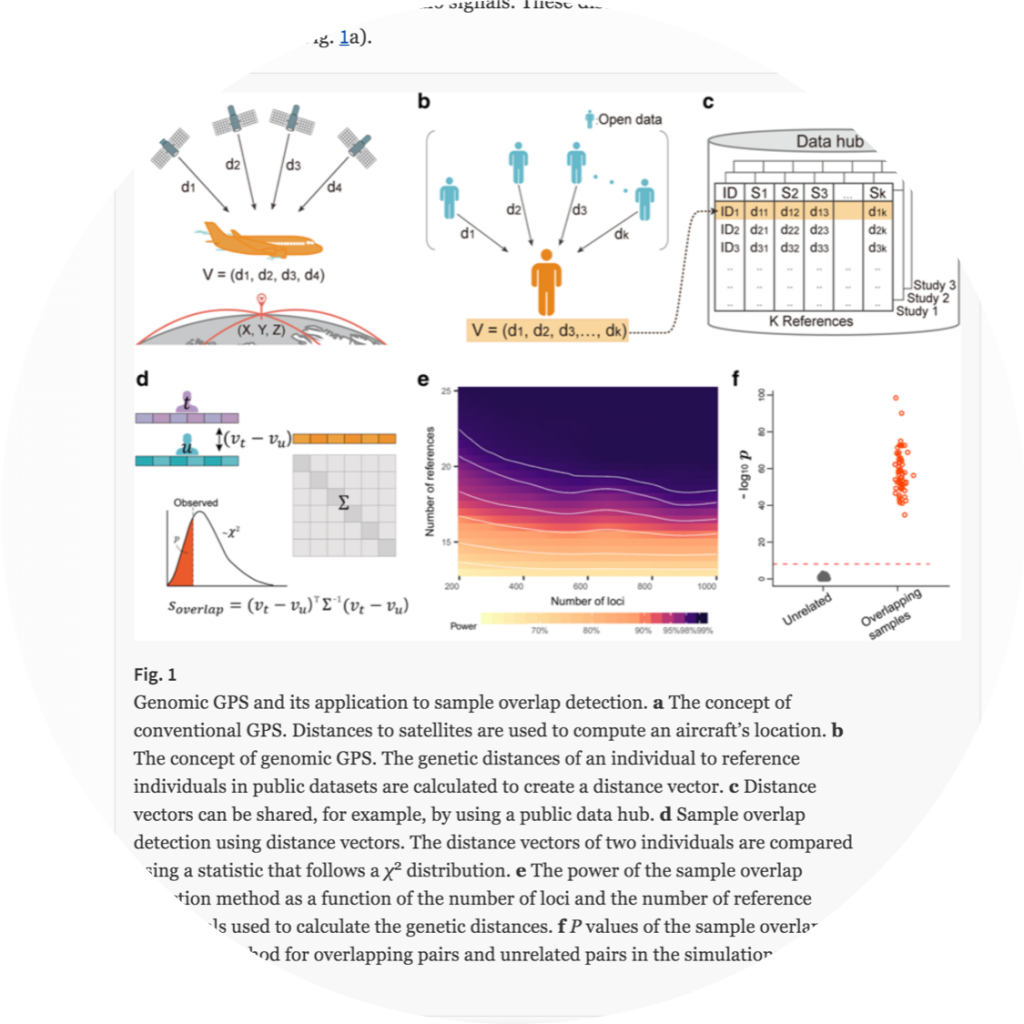
GenomicGPS
Genomic GPS: using genetic distance from individuals to public data for genomic analysis without disclosing personal genomes
- Kunhee Kim, Hyungryul Baik, Chloe Soohyun Jang, Jin Kyung Roh, Eleazer Eskin, Buhm Han. “Genomic GPS: using genetic distance from individuals to public data for genomic analysis without disclosing personal genomes” Genome Biology (2019) 20:175
- Keyword : #privacy, #genetic_distance

RE2C
RE2C is an extended version of the Random Effect model 2 (RE2) implemented in METASOFT
- Cue. H. Lee, Eleazar. Eskin., Buhm Han. “Increasing power of meta-analysis of genome-wide association studies for detecting heterogeneous effects”, Bioinformatics, vol. 33, no. 14m pp. i-379-i388, 2017.
- Keyword : #meta_analysis, #random_effect
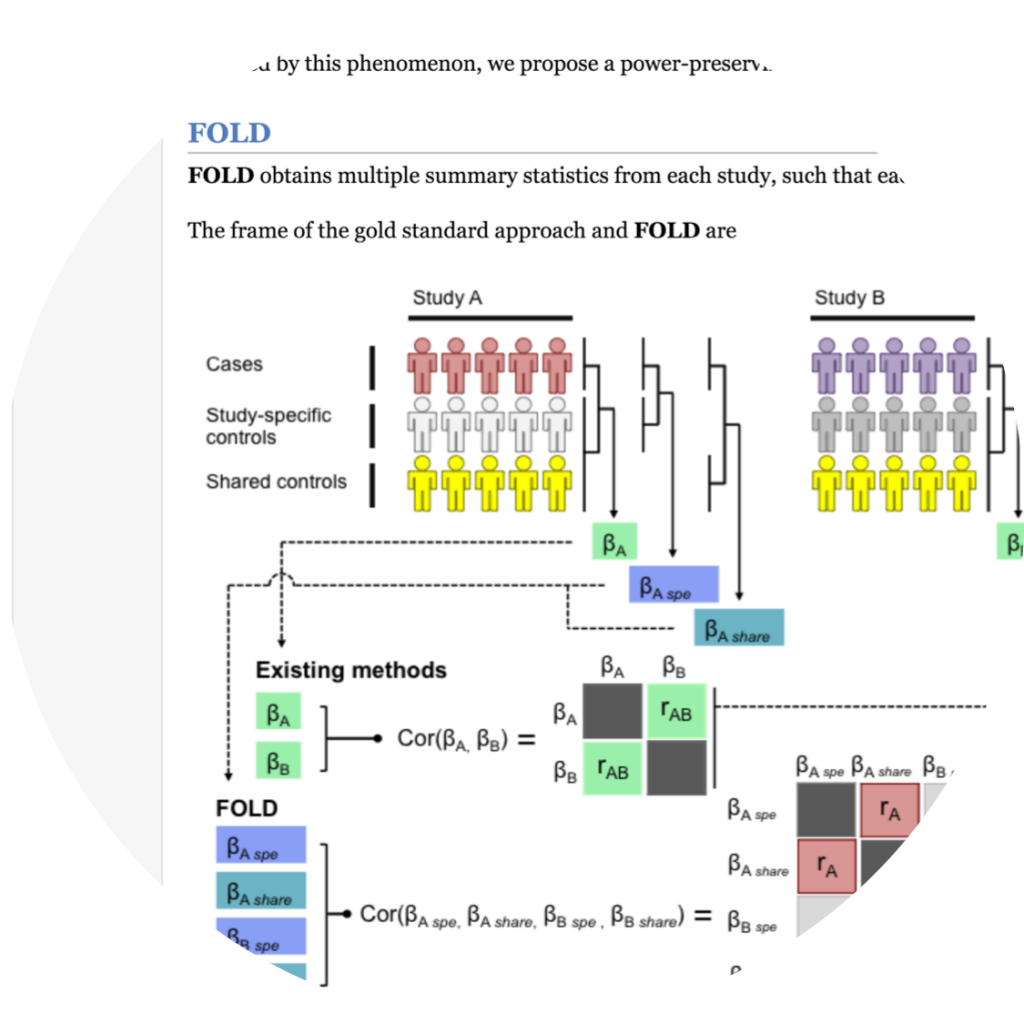
FOLD
FOLD is to obtain a set of summary statistics from each study, where each statistic is calculated using a subset of controls that is homogeneous in terms of their information.
- Eunji Kim, Buhm Han, “A power-preserving meta-analysis framework for overlapping samples”, Bioinformatics. 33(24):3947-3954. 2017
- Keyword : #meta_analysis, #fixed_effect
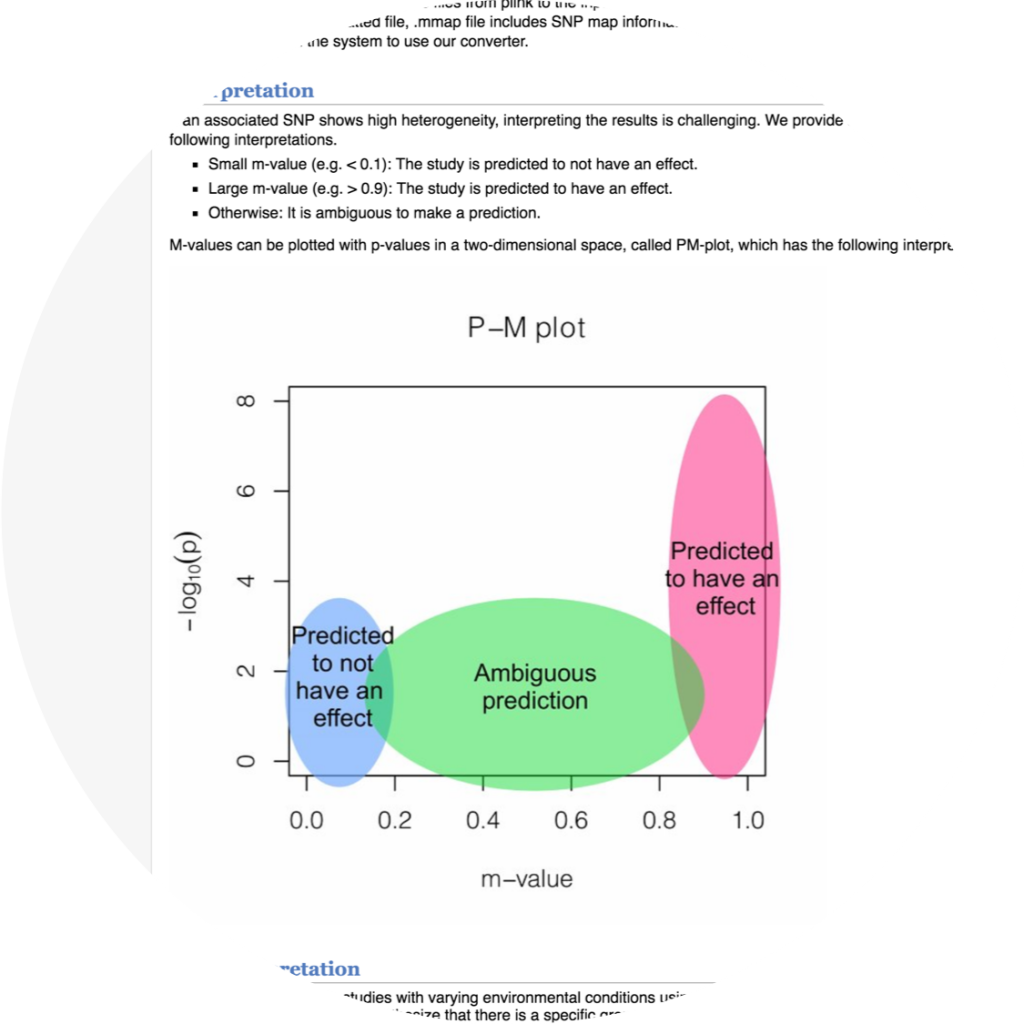
METASOFT
METASOFT performs meta-analysis with new random effects model and calculates m-values (posterior probability that effects will exist).
- Buhm Han, Eleazar Eskin. “Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies.” The American Journal of Human Genetics. 88(5):586-598, 2011.
- Buhm Han, Eleazar Eskin. “Interpreting Meta-Analysis of Genome-Wide Association Studies.” PLoS Genetics. 8(3):e1002555, 2012.
- Keyword : #meta_analysis, #random_effect

SLIDE
SLIDE corrects for multiple testing for millions of markers.
- Buhm Han, Hyun Min Kang, Eleazar Eskin. “Rapid and Accurate Multiple Testing Correction and Power Estimation for Millions of Correlated Markers.” PLoS Genetics. 5:4,e1000456, 2009.
- Keyword : #multiple_testing, #correlation

